Tidy the data
Prerequisites

Here we’ll focus on tidyr, a package that provides a bunch of tools to help tidy up your messy datasets.
suppressWarnings(library(tidyverse))Tidy data

You can represent the same underlying data in multiple ways.
The example below shows the same data organised in four different ways. Each dataset shows the same values of four variables country, year, population, and cases, but each dataset organises the values in a different way.
table1## # A tibble: 6 × 4
## country year cases population
## <chr> <int> <int> <int>
## 1 Afghanistan 1999 745 19987071
## 2 Afghanistan 2000 2666 20595360
## 3 Brazil 1999 37737 172006362
## 4 Brazil 2000 80488 174504898
## 5 China 1999 212258 1272915272
## 6 China 2000 213766 1280428583table2## # A tibble: 12 × 4
## country year type count
## <chr> <int> <chr> <int>
## 1 Afghanistan 1999 cases 745
## 2 Afghanistan 1999 population 19987071
## 3 Afghanistan 2000 cases 2666
## 4 Afghanistan 2000 population 20595360
## 5 Brazil 1999 cases 37737
## 6 Brazil 1999 population 172006362
## 7 Brazil 2000 cases 80488
## 8 Brazil 2000 population 174504898
## 9 China 1999 cases 212258
## 10 China 1999 population 1272915272
## 11 China 2000 cases 213766
## 12 China 2000 population 1280428583table3## # A tibble: 6 × 3
## country year rate
## * <chr> <int> <chr>
## 1 Afghanistan 1999 745/19987071
## 2 Afghanistan 2000 2666/20595360
## 3 Brazil 1999 37737/172006362
## 4 Brazil 2000 80488/174504898
## 5 China 1999 212258/1272915272
## 6 China 2000 213766/1280428583# Spread across two tibbles
table4a # cases## # A tibble: 3 × 3
## country `1999` `2000`
## * <chr> <int> <int>
## 1 Afghanistan 745 2666
## 2 Brazil 37737 80488
## 3 China 212258 213766table4b # population## # A tibble: 3 × 3
## country `1999` `2000`
## * <chr> <int> <int>
## 1 Afghanistan 19987071 20595360
## 2 Brazil 172006362 174504898
## 3 China 1272915272 1280428583These are all representations of the same underlying data, but they are not equally easy to use.
One dataset, the tidy dataset, will be much easier to work with inside the tidyverse.

There are three interrelated rules which make a dataset tidy:
- Each variable must have its own column.
- Each observation must have its own row.
- Each value must have its own cell.
Figure shows the rules visually.

Following three rules makes a dataset tidy: variables are in columns, observations are in rows, and values are in cells.

These three rules are interrelated because it’s impossible to only satisfy two of the three.
In this example, only table1 is tidy. It’s the only
representation where each column is a variable.
Why ensure that your data is tidy?
 There are two main
advantages:
There are two main
advantages:
- There’s a general advantage to picking one consistent way of storing data. If you have a consistent data structure, it’s easier to learn the tools that work with it because they have an underlying uniformity.
- There’s a specific advantage to placing variables in columns because it allows R’s vectorised nature to shine.
dplyr, ggplot2, and all the other packages in the tidyverse are designed to work with tidy data.
Here are a couple of small examples showing how you might work with
table1.
# Compute rate per 10,000
table1 %>%
mutate(rate = cases / population * 10000)## # A tibble: 6 × 5
## country year cases population rate
## <chr> <int> <int> <int> <dbl>
## 1 Afghanistan 1999 745 19987071 0.373
## 2 Afghanistan 2000 2666 20595360 1.29
## 3 Brazil 1999 37737 172006362 2.19
## 4 Brazil 2000 80488 174504898 4.61
## 5 China 1999 212258 1272915272 1.67
## 6 China 2000 213766 1280428583 1.67# Compute cases per year
table1 %>%
count(year, wt = cases)## # A tibble: 2 × 2
## year n
## <int> <int>
## 1 1999 250740
## 2 2000 296920Spreading and gathering

The first step is always to: figure out what the variables and observations are.
The second step is to solve one of two common problems:
One variable might be spread across multiple columns.
One observation might be scattered across multiple rows.
To fix these problems, you’ll need the two most important functions in tidyr:
gather()spread()
gather()
A common problem is a dataset where:
some of the column names are NOT names of variables RATHER observations/values of a variable themselves.
Take table4a: the column names 1999 and
2000 represent values of the year variable,
and each row represents two observations, not one.
table4a## # A tibble: 3 × 3
## country `1999` `2000`
## * <chr> <int> <int>
## 1 Afghanistan 745 2666
## 2 Brazil 37737 80488
## 3 China 212258 213766To tidy a dataset like this, we need to gather those columns into a new pair of variables.
To describe that operation we need three parameters:
The set of columns that represent values, not variables.
The name of the variable whose values form the column names. I call that the
key, and here it isyear.The name of the variable whose values are spread over the cells. I call that
value, and here it’s the number ofcases.
table4a %>%
gather(`1999`, `2000`, key = "year", value = "cases")## # A tibble: 6 × 3
## country year cases
## <chr> <chr> <int>
## 1 Afghanistan 1999 745
## 2 Brazil 1999 37737
## 3 China 1999 212258
## 4 Afghanistan 2000 2666
## 5 Brazil 2000 80488
## 6 China 2000 213766Here there are only two columns, so we list them individually. Note that “1999” and “2000” are non-syntactic names so we have to surround them in backticks.
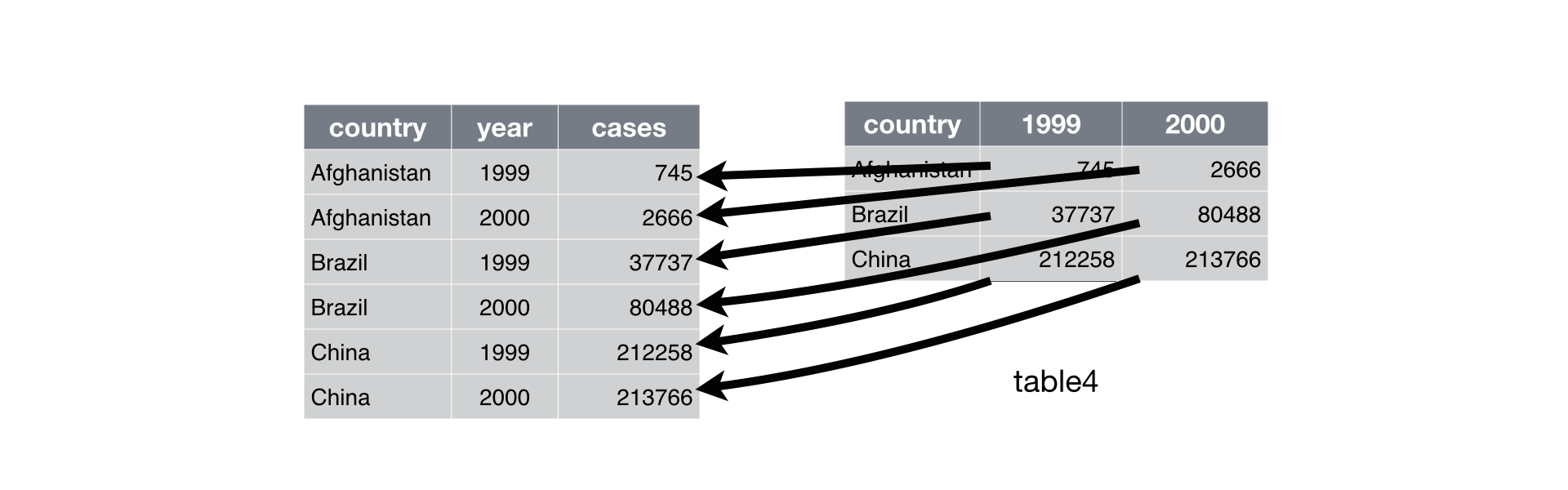
Gathering table4 into a tidy form.
table4b %>%
gather(`1999`, `2000`, key = "year", value = "population")## # A tibble: 6 × 3
## country year population
## <chr> <chr> <int>
## 1 Afghanistan 1999 19987071
## 2 Brazil 1999 172006362
## 3 China 1999 1272915272
## 4 Afghanistan 2000 20595360
## 5 Brazil 2000 174504898
## 6 China 2000 1280428583- To combine the tidied versions of
table4aandtable4binto a single tibble, we need to usedplyr::left_join()
# gather table a
tidy4a <- table4a %>% gather(`1999`, `2000`, key = "year", value = "cases")
# gather table b
tidy4b <- table4b %>% gather(`1999`, `2000`, key = "year", value = "population")
# combine tables once tided
left_join(tidy4a, tidy4b)## Joining, by = c("country", "year")## # A tibble: 6 × 4
## country year cases population
## <chr> <chr> <int> <int>
## 1 Afghanistan 1999 745 19987071
## 2 Brazil 1999 37737 172006362
## 3 China 1999 212258 1272915272
## 4 Afghanistan 2000 2666 20595360
## 5 Brazil 2000 80488 174504898
## 6 China 2000 213766 1280428583spread()
Spreading is the opposite of gathering. Here the issue is that:
an observation is scattered across multiple rows.
For example, take table2: an observation is a country in
a year, but each observation is spread across two rows.
table2## # A tibble: 12 × 4
## country year type count
## <chr> <int> <chr> <int>
## 1 Afghanistan 1999 cases 745
## 2 Afghanistan 1999 population 19987071
## 3 Afghanistan 2000 cases 2666
## 4 Afghanistan 2000 population 20595360
## 5 Brazil 1999 cases 37737
## 6 Brazil 1999 population 172006362
## 7 Brazil 2000 cases 80488
## 8 Brazil 2000 population 174504898
## 9 China 1999 cases 212258
## 10 China 1999 population 1272915272
## 11 China 2000 cases 213766
## 12 China 2000 population 1280428583To tidy this up, we need two parameters:
The column that contains variable names, the
keycolumn.The column that contains values forms multiple variables, the
valuecolumn.
spread(table2, key = type, value = count)## # A tibble: 6 × 4
## country year cases population
## <chr> <int> <int> <int>
## 1 Afghanistan 1999 745 19987071
## 2 Afghanistan 2000 2666 20595360
## 3 Brazil 1999 37737 172006362
## 4 Brazil 2000 80488 174504898
## 5 China 1999 212258 1272915272
## 6 China 2000 213766 1280428583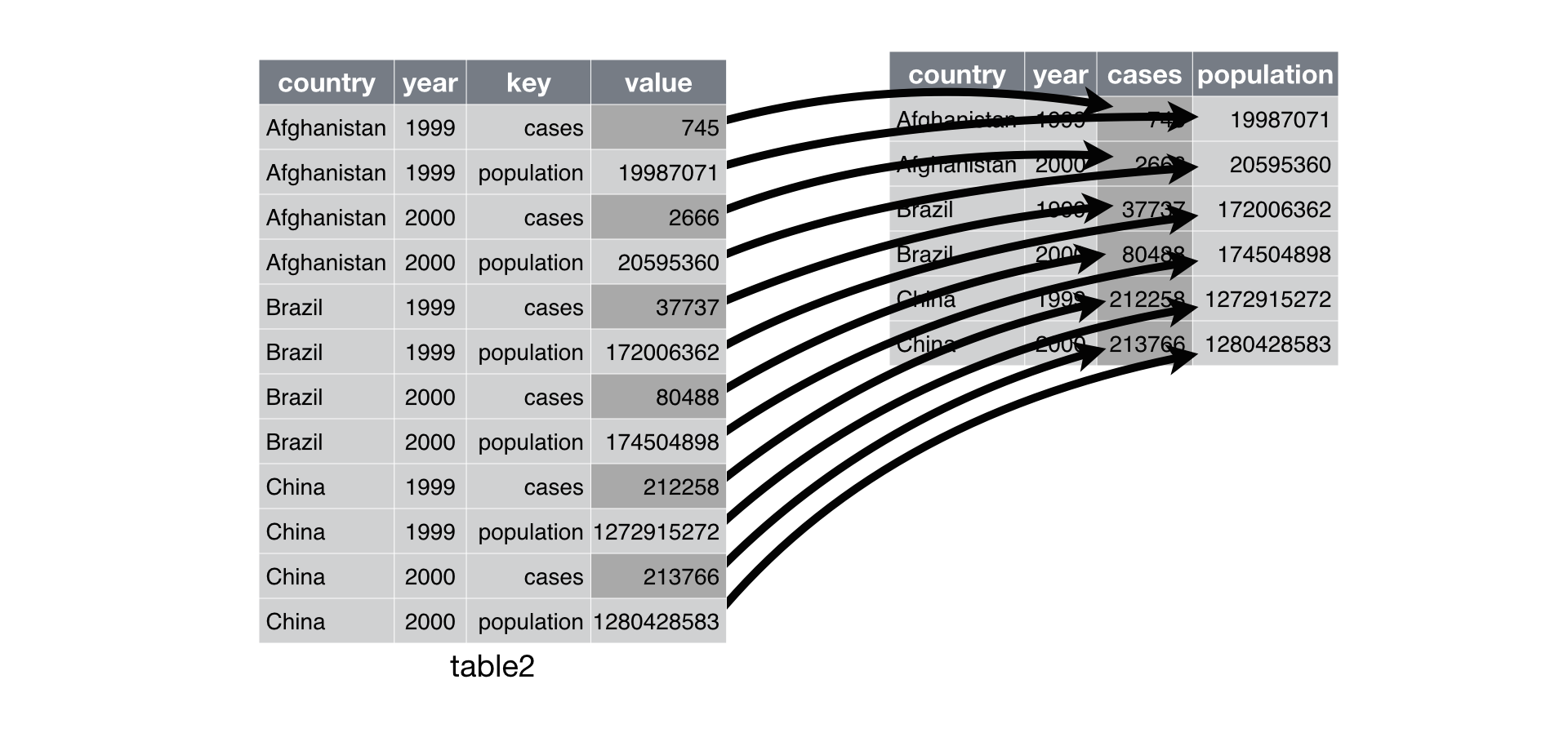
Spreading table2 makes it tidy

gather()makes wide tables narrower and longer
spread()makes long tables shorter and wider
Separating and uniting
table3 has a different problem :
one column (
rate) that contains two variables (casesandpopulation)
table3## # A tibble: 6 × 3
## country year rate
## * <chr> <int> <chr>
## 1 Afghanistan 1999 745/19987071
## 2 Afghanistan 2000 2666/20595360
## 3 Brazil 1999 37737/172006362
## 4 Brazil 2000 80488/174504898
## 5 China 1999 212258/1272915272
## 6 China 2000 213766/1280428583To fix this problem, we will need the separate()
function.
The complement of separate() is unite(), to
use when a single variable is spread across multiple columns.
separate()
This function pulls apart one column into multiple columns, by
splitting wherever a separator character appears. Take
table3:
table3## # A tibble: 6 × 3
## country year rate
## * <chr> <int> <chr>
## 1 Afghanistan 1999 745/19987071
## 2 Afghanistan 2000 2666/20595360
## 3 Brazil 1999 37737/172006362
## 4 Brazil 2000 80488/174504898
## 5 China 1999 212258/1272915272
## 6 China 2000 213766/1280428583The rate column contains both cases and
population variables, and we need to split it into two
variables.
separate() takes the name of the column to separate, and
the names of the columns to separate into, as shown in Figure
@ref(fig:tidy-separate) and the code below.
table3 %>%
separate(rate, into = c("cases", "population"))## # A tibble: 6 × 4
## country year cases population
## <chr> <int> <chr> <chr>
## 1 Afghanistan 1999 745 19987071
## 2 Afghanistan 2000 2666 20595360
## 3 Brazil 1999 37737 172006362
## 4 Brazil 2000 80488 174504898
## 5 China 1999 212258 1272915272
## 6 China 2000 213766 1280428583
Separating table3 makes it tidy

By default,
separate()will split values wherever it sees a non-alphanumeric character.
table3 %>%
separate(rate, into = c("cases", "population"), sep = "/")Look carefully at the column types: you’ll notice that
case and population are character columns.
This is the default behaviour in separate(): it leaves the
type of the column as is. Here, however, it’s not very useful as those
really are numbers. We can ask separate() to try and
convert to better types using convert = TRUE:
table3 %>%
separate(rate, into = c("cases", "population"), convert = TRUE)## # A tibble: 6 × 4
## country year cases population
## <chr> <int> <int> <int>
## 1 Afghanistan 1999 745 19987071
## 2 Afghanistan 2000 2666 20595360
## 3 Brazil 1999 37737 172006362
## 4 Brazil 2000 80488 174504898
## 5 China 1999 212258 1272915272
## 6 China 2000 213766 1280428583You can also pass a vector of integers to sep.
separate() will interpret the integers as positions to
split at. Positive values start at 1 on the far-left of the strings;
negative value start at -1 on the far-right of the strings. When using
integers to separate strings, the length of sep should be
one less than the number of names in into.
table3 %>%
separate(year, into = c("century", "year"), sep = 2)## # A tibble: 6 × 4
## country century year rate
## <chr> <chr> <chr> <chr>
## 1 Afghanistan 19 99 745/19987071
## 2 Afghanistan 20 00 2666/20595360
## 3 Brazil 19 99 37737/172006362
## 4 Brazil 20 00 80488/174504898
## 5 China 19 99 212258/1272915272
## 6 China 20 00 213766/1280428583unite()
This function is the inverse of separate(): it combines
multiple columns into a single column.

Uniting table5 makes it tidy
We can use unite() to rejoin the century and
year columns that we created in the last example. That data is
saved as tidyr::table5.
table5 %>%
unite(new, century, year)## # A tibble: 6 × 3
## country new rate
## <chr> <chr> <chr>
## 1 Afghanistan 19_99 745/19987071
## 2 Afghanistan 20_00 2666/20595360
## 3 Brazil 19_99 37737/172006362
## 4 Brazil 20_00 80488/174504898
## 5 China 19_99 212258/1272915272
## 6 China 20_00 213766/1280428583In this case we also need to use the sep argument.
The default will place an underscore (_) between the
values from different columns.
Here we don’t want any separator so we use "":
table5 %>%
unite(new, century, year, sep = "")## # A tibble: 6 × 3
## country new rate
## <chr> <chr> <chr>
## 1 Afghanistan 1999 745/19987071
## 2 Afghanistan 2000 2666/20595360
## 3 Brazil 1999 37737/172006362
## 4 Brazil 2000 80488/174504898
## 5 China 1999 212258/1272915272
## 6 China 2000 213766/1280428583Missing values
Changing the representation of a dataset brings up an important subtlety of missing values.
A value can be missing in one of two possible ways:
Explicitly –> flagged with
NA.Implicitly –> simply not present in the data.
Let’s illustrate this idea with a very simple data set:
stocks <- tibble(
year = c(2015, 2015, 2015, 2015, 2016, 2016, 2016),
qtr = c( 1, 2, 3, 4, 2, 3, 4),
return = c(1.88, 0.59, 0.35, NA, 0.92, 0.17, 2.66)
)
print(stocks)## # A tibble: 7 × 3
## year qtr return
## <dbl> <dbl> <dbl>
## 1 2015 1 1.88
## 2 2015 2 0.59
## 3 2015 3 0.35
## 4 2015 4 NA
## 5 2016 2 0.92
## 6 2016 3 0.17
## 7 2016 4 2.66There are two missing values in this dataset.
We can make the implicit missing value explicit by putting years in the columns:
stocks %>%
spread(year, return)## # A tibble: 4 × 3
## qtr `2015` `2016`
## <dbl> <dbl> <dbl>
## 1 1 1.88 NA
## 2 2 0.59 0.92
## 3 3 0.35 0.17
## 4 4 NA 2.66Because these explicit missing values may not be important in other
representations of the data, you can set na.rm = TRUE in
gather() to turn explicit missing values implicit:
stocks %>%
spread(year, return) %>%
gather(year, return, `2015`:`2016`, na.rm = TRUE)## # A tibble: 6 × 3
## qtr year return
## <dbl> <chr> <dbl>
## 1 1 2015 1.88
## 2 2 2015 0.59
## 3 3 2015 0.35
## 4 2 2016 0.92
## 5 3 2016 0.17
## 6 4 2016 2.66Another important tool for making missing values explicit in tidy
data is complete():
stocks %>%
complete(year, qtr)## # A tibble: 8 × 3
## year qtr return
## <dbl> <dbl> <dbl>
## 1 2015 1 1.88
## 2 2015 2 0.59
## 3 2015 3 0.35
## 4 2015 4 NA
## 5 2016 1 NA
## 6 2016 2 0.92
## 7 2016 3 0.17
## 8 2016 4 2.66
complete()takes a set of columns, and finds all unique combinations.
It then ensures the original dataset contains all those values,
filling in explicit NAs where necessary.
Sometimes when a data source has primarily been used for data entry, missing values indicate that the previous value should be carried forward:
treatment <- tribble(
~ person, ~ treatment, ~response,
"Derrick Whitmore", 1, 7,
NA, 2, 10,
NA, 3, 9,
"Katherine Burke", 1, 4
)You can fill in these missing values with fill(). It
takes a set of columns where you want missing values to be replaced by
the most recent non-missing value (sometimes called last observation
carried forward).
treatment %>%
fill(person)## # A tibble: 4 × 3
## person treatment response
## <chr> <dbl> <dbl>
## 1 Derrick Whitmore 1 7
## 2 Derrick Whitmore 2 10
## 3 Derrick Whitmore 3 9
## 4 Katherine Burke 1 4Case Study
The tidyr::who dataset contains tuberculosis (TB)
cases broken down by year, country, age, gender, and diagnosis
method.
The data comes from the 2014 World Health Organization Global Tuberculosis Report, available at http://www.who.int/tb/country/data/download/en/.
print(who, n=4)## # A tibble: 7,240 × 60
## country iso2 iso3 year new_sp_m014 new_sp_m1524 new_sp_m2534 new_sp_m3544 new_sp_m4554
## <chr> <chr> <chr> <int> <int> <int> <int> <int> <int>
## 1 Afghan… AF AFG 1980 NA NA NA NA NA
## 2 Afghan… AF AFG 1981 NA NA NA NA NA
## 3 Afghan… AF AFG 1982 NA NA NA NA NA
## 4 Afghan… AF AFG 1983 NA NA NA NA NA
## # … with 7,236 more rows, and 51 more variables: new_sp_m5564 <int>, new_sp_m65 <int>,
## # new_sp_f014 <int>, new_sp_f1524 <int>, new_sp_f2534 <int>, new_sp_f3544 <int>,
## # new_sp_f4554 <int>, new_sp_f5564 <int>, new_sp_f65 <int>, new_sn_m014 <int>,
## # new_sn_m1524 <int>, new_sn_m2534 <int>, new_sn_m3544 <int>, new_sn_m4554 <int>,
## # new_sn_m5564 <int>, new_sn_m65 <int>, new_sn_f014 <int>, new_sn_f1524 <int>,
## # new_sn_f2534 <int>, new_sn_f3544 <int>, new_sn_f4554 <int>, new_sn_f5564 <int>,
## # new_sn_f65 <int>, new_ep_m014 <int>, new_ep_m1524 <int>, new_ep_m2534 <int>, …It contains redundant columns, odd variable codes, and many missing values.
country,iso2, andiso3are three variables that redundantly specify the country.yearis clearly also a variable.We don’t know what all the other columns are but given the structure in the variable names (e.g.
new_sp_m014,new_ep_m014,new_ep_f014) these are likely to be values, not variables.
So we need to gather() together all the columns from
new_sp_m014 to newrel_f65.
We don’t know what those values represent yet, so we’ll give them the
generic name "key".
We know the cells represent the count of cases, so we’ll use the
variable cases.
There are a lot of missing values in the current representation, so
for now we’ll use na.rm just so we can focus on the values
that are present.
who1 <- who %>%
gather(new_sp_m014:newrel_f65, key = "key", value = "cases", na.rm = TRUE)
who1## # A tibble: 76,046 × 6
## country iso2 iso3 year key cases
## <chr> <chr> <chr> <int> <chr> <int>
## 1 Afghanistan AF AFG 1997 new_sp_m014 0
## 2 Afghanistan AF AFG 1998 new_sp_m014 30
## 3 Afghanistan AF AFG 1999 new_sp_m014 8
## 4 Afghanistan AF AFG 2000 new_sp_m014 52
## 5 Afghanistan AF AFG 2001 new_sp_m014 129
## 6 Afghanistan AF AFG 2002 new_sp_m014 90
## 7 Afghanistan AF AFG 2003 new_sp_m014 127
## 8 Afghanistan AF AFG 2004 new_sp_m014 139
## 9 Afghanistan AF AFG 2005 new_sp_m014 151
## 10 Afghanistan AF AFG 2006 new_sp_m014 193
## # … with 76,036 more rowsWe can get some hint of the structure of the values in the new
key column by counting them:
who1 %>%
count(key)## # A tibble: 56 × 2
## key n
## <chr> <int>
## 1 new_ep_f014 1032
## 2 new_ep_f1524 1021
## 3 new_ep_f2534 1021
## 4 new_ep_f3544 1021
## 5 new_ep_f4554 1017
## 6 new_ep_f5564 1017
## 7 new_ep_f65 1014
## 8 new_ep_m014 1038
## 9 new_ep_m1524 1026
## 10 new_ep_m2534 1020
## # … with 46 more rowsIt tells us:
The first three letters of each column denote whether the column contains new or old cases of TB. In this dataset, each column contains new cases.
The next two letters describe the type of TB:
relstands for cases of relapseepstands for cases of extrapulmonary TBsnstands for cases of pulmonary TB that could not be diagnosed by a pulmonary smear (smear negative)spstands for cases of pulmonary TB that could be diagnosed be a pulmonary smear (smear positive)
The sixth letter gives the sex of TB patients. The dataset groups cases by males (
m) and females (f).The remaining numbers gives the age group. The dataset groups cases into seven age groups:
014= 0 – 14 years old1524= 15 – 24 years old2534= 25 – 34 years old- …
65= 65 or older
We need to make a minor fix to the format of the column names.
who2 <- who1 %>%
mutate(key = stringr::str_replace(key, "newrel", "new_rel"))
who2## # A tibble: 76,046 × 6
## country iso2 iso3 year key cases
## <chr> <chr> <chr> <int> <chr> <int>
## 1 Afghanistan AF AFG 1997 new_sp_m014 0
## 2 Afghanistan AF AFG 1998 new_sp_m014 30
## 3 Afghanistan AF AFG 1999 new_sp_m014 8
## 4 Afghanistan AF AFG 2000 new_sp_m014 52
## 5 Afghanistan AF AFG 2001 new_sp_m014 129
## 6 Afghanistan AF AFG 2002 new_sp_m014 90
## 7 Afghanistan AF AFG 2003 new_sp_m014 127
## 8 Afghanistan AF AFG 2004 new_sp_m014 139
## 9 Afghanistan AF AFG 2005 new_sp_m014 151
## 10 Afghanistan AF AFG 2006 new_sp_m014 193
## # … with 76,036 more rowsWe can separate the values in each code with two passes of
separate().
who3 <- who2 %>%
separate(key, c("new", "type", "sexage"), sep = "_")
who3## # A tibble: 76,046 × 8
## country iso2 iso3 year new type sexage cases
## <chr> <chr> <chr> <int> <chr> <chr> <chr> <int>
## 1 Afghanistan AF AFG 1997 new sp m014 0
## 2 Afghanistan AF AFG 1998 new sp m014 30
## 3 Afghanistan AF AFG 1999 new sp m014 8
## 4 Afghanistan AF AFG 2000 new sp m014 52
## 5 Afghanistan AF AFG 2001 new sp m014 129
## 6 Afghanistan AF AFG 2002 new sp m014 90
## 7 Afghanistan AF AFG 2003 new sp m014 127
## 8 Afghanistan AF AFG 2004 new sp m014 139
## 9 Afghanistan AF AFG 2005 new sp m014 151
## 10 Afghanistan AF AFG 2006 new sp m014 193
## # … with 76,036 more rowsThen we might as well drop the new column because it’s
constant in this dataset. While we’re dropping columns, let’s also drop
iso2 and iso3 since they’re redundant.
who3 %>%
count(new)## # A tibble: 1 × 2
## new n
## <chr> <int>
## 1 new 76046who4 <- who3 %>%
select(-new, -iso2, -iso3)Next we’ll separate sexage into sex and
age by splitting after the first character:
who5 <- who4 %>%
separate(sexage, c("sex", "age"), sep = 1)
print(who5, n=5)## # A tibble: 76,046 × 6
## country year type sex age cases
## <chr> <int> <chr> <chr> <chr> <int>
## 1 Afghanistan 1997 sp m 014 0
## 2 Afghanistan 1998 sp m 014 30
## 3 Afghanistan 1999 sp m 014 8
## 4 Afghanistan 2000 sp m 014 52
## 5 Afghanistan 2001 sp m 014 129
## # … with 76,041 more rowsA work by Matteo Cereda and Fabio Iannelli